Using the merge() function in R on big tables can be time consuming. Luckily the join functions in the new package dplyr are much faster. The package offers four different joins:
inner_join(similar to merge withall.x=Fandall.y=F)left_join(similar to merge withall.x=Tandall.y=F)semi_join(not really an equivalent inmerge()unless y only includes join fields)anti_join(no equivalent inmerge(), this is all x without a match in y)
- Example 1: Combine Data by Two ID Columns Using merge Function. In Example 1, I'll illustrate how to apply the merge function to combine data frames based on multiple ID columns. For this, we have to specify the by argument of the merge function to be equal to a vector of ID column names (i.e. By = c('ID1', 'ID2')).
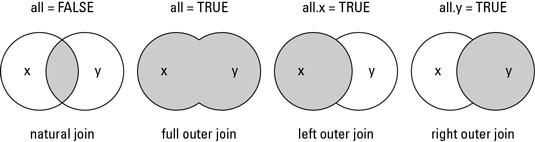
- Example 5: semijoin dplyr R Function The four previous join functions (i.e. Innerjoin, leftjoin, rightjoin, and fulljoin) are so called mutating joins. Mutating joins combine variables from the two data sources. The next two join functions (i.e. Semijoin and antijoin) are so called filtering joins.
- A left join in R is a merge operation between two data frames where the merge returns all of the rows from one table (the left side) and any matching rows from the second table. A left join in R will NOT return values of the second table which do not already exist in the first table.
I can't find a great discussion of the advantages of the dplyr join functions but I do see a help response from Hadley Wickham, dplyr's creator, here that briefly lists the advantages:
A character vector of variables to join. If NULL, the default,.join will do a natural join, using all variables with common names across the two tables. A message lists the variables so that you can check they're right (to suppress the message, simply explicitly list the variables that you want to join).
- rows are kept in existing order
- much faster
- tells you what keys you're merging by (if you don't supply)
- also work with database tables.
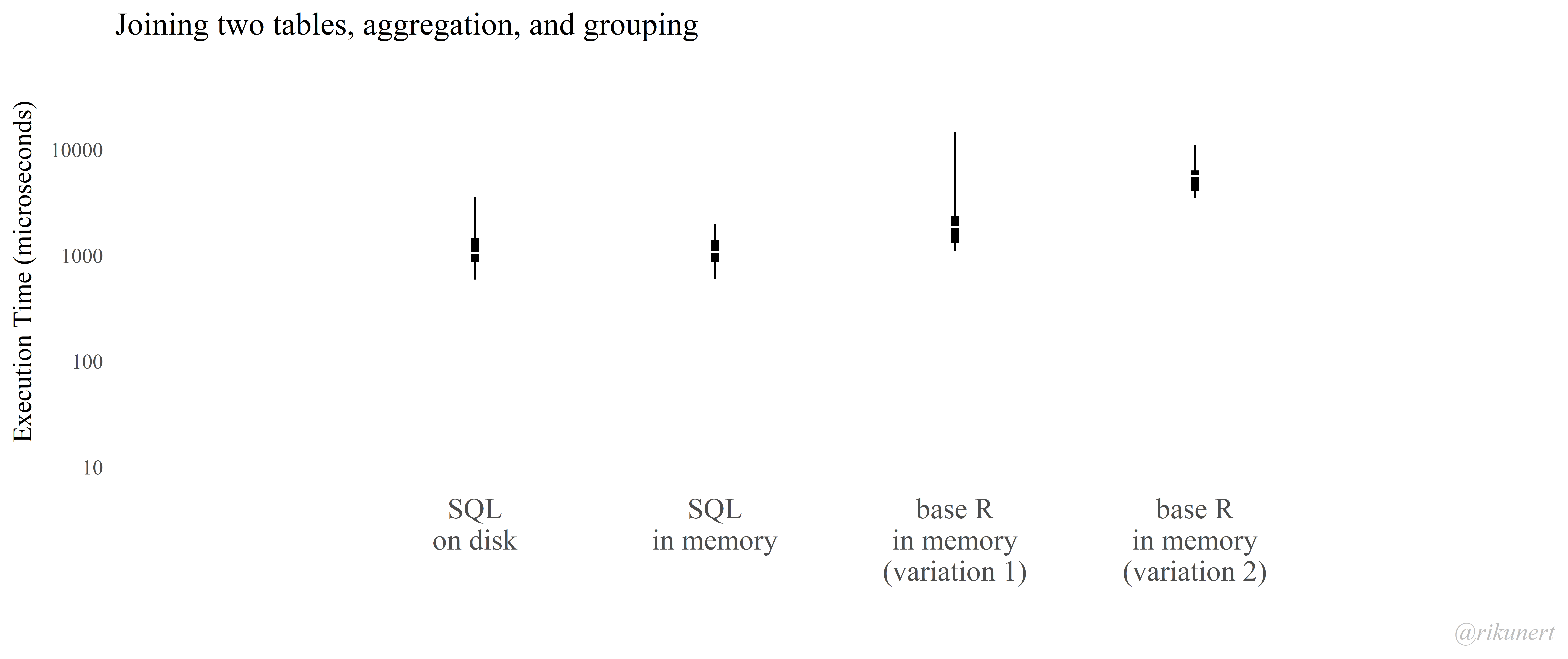
Of course, the advantage that matters most is the speed. In this tiny example using a table with more than 6 million records the inner_join function is 43 times faster!
Adding Columns
To merge two data frames (datasets) horizontally, use the merge function. In most cases, you join two data frames by one or more common key variables (i.e., an inner join).
# merge two data frames by ID
total <- merge(data frameA,data frameB,by='ID')
# merge two data frames by ID and Country
total <- merge(data frameA,data frameB,by=c('ID','Country'))
Adding Rows
To join two data frames (datasets) vertically, use the rbind function. The two data frames must have the same variables, but they do not have to be in the same order.
total <- rbind(data frameA, data frameB)
If data frameA has variables that data frameB does not, then either:
Full Join Function In R
- Delete the extra variables in data frameA or
- Create the additional variables in data frameB and set them to NA (missing)
before joining them with rbind( ).
Going Further
Left Join Function In R

inner_join(similar to merge withall.x=Fandall.y=F)left_join(similar to merge withall.x=Tandall.y=F)semi_join(not really an equivalent inmerge()unless y only includes join fields)anti_join(no equivalent inmerge(), this is all x without a match in y)
- Example 1: Combine Data by Two ID Columns Using merge Function. In Example 1, I'll illustrate how to apply the merge function to combine data frames based on multiple ID columns. For this, we have to specify the by argument of the merge function to be equal to a vector of ID column names (i.e. By = c('ID1', 'ID2')).
- Example 5: semijoin dplyr R Function The four previous join functions (i.e. Innerjoin, leftjoin, rightjoin, and fulljoin) are so called mutating joins. Mutating joins combine variables from the two data sources. The next two join functions (i.e. Semijoin and antijoin) are so called filtering joins.
- A left join in R is a merge operation between two data frames where the merge returns all of the rows from one table (the left side) and any matching rows from the second table. A left join in R will NOT return values of the second table which do not already exist in the first table.
I can't find a great discussion of the advantages of the dplyr join functions but I do see a help response from Hadley Wickham, dplyr's creator, here that briefly lists the advantages:
A character vector of variables to join. If NULL, the default,.join will do a natural join, using all variables with common names across the two tables. A message lists the variables so that you can check they're right (to suppress the message, simply explicitly list the variables that you want to join).
- rows are kept in existing order
- much faster
- tells you what keys you're merging by (if you don't supply)
- also work with database tables.
Of course, the advantage that matters most is the speed. In this tiny example using a table with more than 6 million records the inner_join function is 43 times faster!
Adding Columns
To merge two data frames (datasets) horizontally, use the merge function. In most cases, you join two data frames by one or more common key variables (i.e., an inner join).
# merge two data frames by ID
total <- merge(data frameA,data frameB,by='ID')
# merge two data frames by ID and Country
total <- merge(data frameA,data frameB,by=c('ID','Country'))
Adding Rows
To join two data frames (datasets) vertically, use the rbind function. The two data frames must have the same variables, but they do not have to be in the same order.
total <- rbind(data frameA, data frameB)
If data frameA has variables that data frameB does not, then either:
Full Join Function In R
- Delete the extra variables in data frameA or
- Create the additional variables in data frameB and set them to NA (missing)
before joining them with rbind( ).
Going Further
Left Join Function In R
To practice manipulating data frames with the dplyr package, try this interactive course on data frame manipulation in R.

